Niamh Tumelty, Head of Open Research Services, Cambridge University Libraries
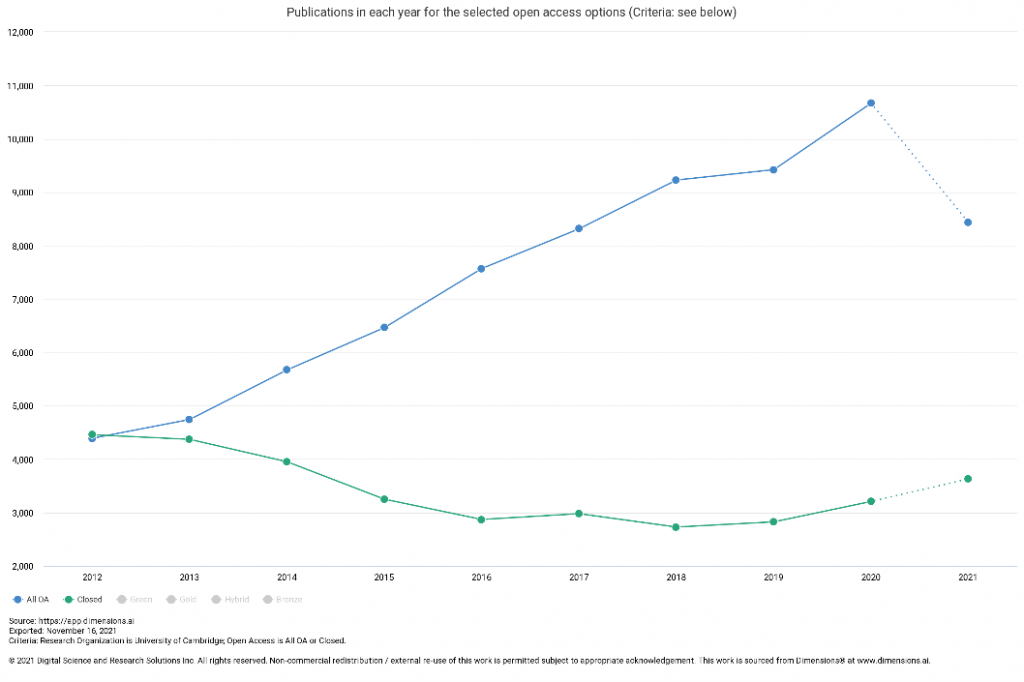
Yesterday it was announced that the White House Office of Science and Technology Policy has updated US policy guidance to make the results of taxpayer-supported research immediately available to the American public at no cost:
https://www.whitehouse.gov/ostp/news-updates/2022/08/25/ostp-issues-guidance-to-make-federally-funded-research-freely-available-without-delay/
Federal agencies have been asked to update their public access policies to make publications and supporting data publicly accessible without an embargo. This applies to all federal agencies (the previous policy only applied to those with more than $100 million in annual research and development expenditure) and allows for flexibility for the agencies to decide on some of the details while encouraging alignment of approaches. It applies to all peer-reviewed research articles in journals and includes the potential to also include peer-reviewed book chapters, editorials and peer-reviewed conference proceedings.
The emphasis on “measures to reduce inequities of, and access to, federally funded research and data” is particularly important in light of the serious risk that we will just move from a broken system with built-in inequities around access to information to a new broken system with built-in inequities around whose voices can be heard. Active engagement will be needed to ensure that the agencies take these issues into account and are not contributing to these inequities.
While there will be a time lag in terms of development/updating and implementation of agency policies and we don’t yet have the fine print around licences etc, this will bring requirements for US researchers more closely in line with what many of our researchers already need to do as a result of e.g. UKRI and Wellcome Trust policies. Closer alignment should help address some of the collaborator issues that have arisen following the recent cOAlition S policy updates – though of course a lot will depend on the detail of what each agency puts in place. Researchers availing of US federal funding need to engage now if they would like to influence the approach taken by those who fund their work.
There continues to be a very real question around sustainable business models both from publisher and institutional perspectives, alongside the other big questions around whether the current approaches to scholarly publishing are serving the needs of researchers adequately. It is essential that this doesn’t just become an additional cost for researchers or institutions as many of those who have commented in the past 24 hours fear. Many alternatives to the APC and transitional agreement/big deal approaches have been proposed, from diamond approaches through to completely reimagined approaches to publishing (e.g. Octopus).
There will be mixed feelings about this. While there is likely to be little sympathy for the publishers with the widest profit margins, this move is sure to push more of the smaller publishers, including many (but not all!) learned societies, to think differently. We need to ensure that we understand what researchers most value about these publishers and how to preserve those aspects in whatever comes in future – I am reminded of the thought-provoking comments from our recent working group on open research in the humanities on this topic.
These are big conversations that were already underway and will now take on greater urgency. The greatest challenge of all remains how to change the research culture such researchers can have confidence in sharing their work and expertise in ways that maximise access to their work while also aligning with their (differing!) values and priorities.